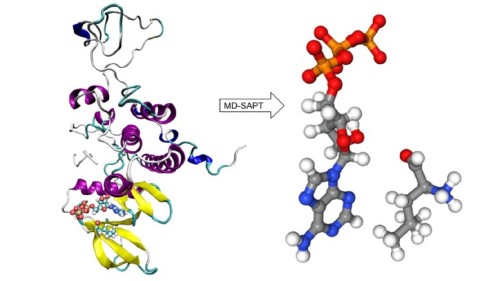
Ashley McDonald of California Polytechnic State University will share her research on MDSAPT, an open-source molecular dynamics software tool that enables calculation of the interaction energy using electronic structure methods.
Noncovalent interactions play an important role in stabilizing small molecule binding events in macromolecular structures, such as a ligand binding to a protein structure. Often, once the residues involved in stabilizing interactions have been identified, a detailed analysis of the molecular interactions is desired, requiring the use of correlated electronic structure methods. In this presentation, we discuss MDSAPT, an open-source molecular dynamics software tool that enables calculation of the interaction energy, using electronic structure methods, between specified residues over the course of the whole simulation. The tool combines trajectory analysis, structure preparation, and quantum chemistry calculations to analyze how interactions change as the structure changes. The tool is implemented as a python package, which can be run as a standalone program from the command line or in jupyter notebook or imported as a python library into a more complex workflow.
The seminar will also discuss McDonald’s work with the Molecular Sciences Software Institute and their resources for teaching best practices in software development.
Join us on Friday, Jan. 26, for this exciting presentation from Ashley McDonald. The presentation will begin at 4:10 p.m. in Hayes Hall 109. We hope to see you there!